Abstract
Introduction Up to one-third of newly diagnosed diffuse large B-cell lymphoma (DLBCL) patients relapse or fail to achieve complete remission following first-line treatment. Adding PET radiomics features to currently used clinical predictors may improve the identification of poor prognosis patients. The objective of this study was to externally validate the radiomics model developed in the HOVON-84 trial (Eertink et al, EJNMMI 2021) using datasets from 6 other DLBCL research studies within the PETRA database.
Methods 1195 newly diagnosed DLBCL patients with baseline 18F-FDG PET/CT scans with 2 years follow up in 7 studies in the PETRA database (https://petralymphoma.org) were included. 308 patients from the HOVON 84 were used as test set previously described. 887 patients from 6 studies in the PETRA database were used as external validation datasets. Primary outcome measures were 2-year progression free survival (PFS) and 2-year time to progression (TTP). Lesions were delineated using a fully automated preselection of 18F-FDG avid structures defined by a standardized Uptake Value (SUV) ≥ 4.0 and volume >3mL. Missed lesions were manually added and non-tumor regions were removed using the ACCURATE tool. Metabolic tumor volume (MTV), the maximum distance between the largest lesion and any other lesion (DmaxBulk) and the peak standardized uptake value (SUVpeak) were extracted for all patients. We tested the predictive value of the currently used international prognostic index (IPI) score and the radiomics model developed in the HOVON-84 trial, which included MTV, DmaxBulk, SUVpeak, WHO performance status and age. Model performance was assessed using the receiver-operator-characteristics area under the curve (AUC). To allow comparison of the continuous radiomics model with individual predicted probabilities per patient with the categorical IPI risk score, the high-risk group for the radiomics model was set as equal the size of the high-risk IPI group. Diagnostic performance was assessed using the positive predictive value (PPV). Differences between model performances of prediction models, expressed as AUC, were assessed with the two-sided DeLong test.
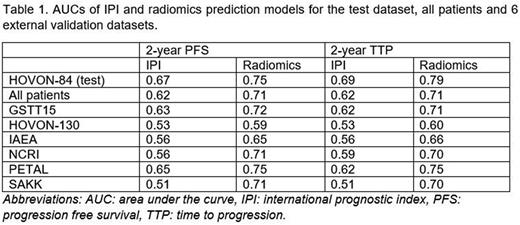
Results Using 2-year PFS as outcome, the IPI model yielded an AUC of 0.62 using all 1195 patients (Table 1). Within the 6 external datasets, the AUC of the IPI model ranged from 0.51 for the SAKK study to 0.65 for the PETAL study. The radiomics model yielded an AUC of 0.71, which was significantly higher than the IPI model (p < 0.001). The AUC of the radiomics model ranged between 0.59 for the HOVON-130 study to 0.75 for the PETAL study. For all external datasets, the AUC of the radiomics model was higher than the AUC of the IPI model. Comparable results were found using 2-year TTP as outcome. The IPI model yielded an AUC of 0.62 and the radiomics model yielded an AUC of 0.71 when using all 1195 patients (p < 0.001). Again, the AUCs of the radiomics models were consistently higher than the AUCs of the IPI model for all individual studies.
High-risk patients according to the IPI model had a 2-year PFS of 60.3% (95% confidence interval (CI): 54.4-67.0). High risk patients by the radiomics model had a 2-year PFS of 50.9% (95% CI: 44.8-57.7). The PPV increased from 35.5% for the IPI model to 49.1% for the radiomics model. For 2-year TTP as outcome, high-risk IPI patients had an overall survival rate of 66.4% (95%CI: 60.3-73.0). High-risk radiomics patients had a survival rate of 55.5% (95% CI: 49.1-62.6). The PPV increased from 33.7% to 44.6% for the radiomics model compared to the IPI model.
Conclusion The radiomics model that was developed in the HOVON-84 dataset remained predictive of outcome in 6 independent first-line DLBCL studies, and had higher model performance than the currently used IPI risk score in all studies. Differences between IPI and radiomics model performance between individual studies are most likely caused by differences in patient characteristics between studies. Our radiomics model is able to select high-risk patients more accurately than the IPI model, with higher model performance and PPV.
Disclosures
Lugtenburg:Celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Travel Grants; Roche: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Genmab: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Travel Grants, Research Funding; AbbVie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Incyte: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Regeneron: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Servier: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Barrington:Bristol Myers Squibb international: Research Funding; Astra Zeneca: Research Funding; Amgen Ltd: Research Funding; Novartis: Research Funding; Pfizer: Research Funding; Takeda: Research Funding. Zucca:Mei Pharma: Other: advisory board fees ; Astra Zeneca: Other: advisory board fees ; Celltrion Healthcare: Other: advisory board fees ; Abbvie: Other: travel grant; Gilead: Other: travel grant, expert statements; Bristol-Myers Squibb: Other: expert statements; MSD: Other: expert statements; Incyte: Other: advisory board fee, Research Funding; BeiGene: Other: advisory board fee, Research Funding; Janssen: Research Funding; Roche: Research Funding; Celgene: Other: advisory board fees, Research Funding; Miltenyi Biomedicine: Other: advisory board fee. Chamuleau:Gilead: Research Funding; Roche: Honoraria; BMS/Celgene: Honoraria, Research Funding; Genmab: Research Funding; Abbvie: Honoraria; Novartis: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.